'Group by' for batch processing - parent child app
Hi Forum,
I'm trying to set up a Parent-Child batch analysis of a set of images. The images consist of multiple positions with every position having 2 channels (parent and child channel). Attached is the general naming of the files - d0 (parent) and d1 (child) are the two channels. A01 and f00 are identifiers of different positions.
I've tried grouping them accordingly using this syntax : [A-A]+(\d\d)+.+(\d\d). However it does not separate images with different numbers after 'f'. It should have 2 images per group.
Also, as I haven't been able to try this, would this even be the way to batch process images for the parent-child app?
Thank you,
Henri
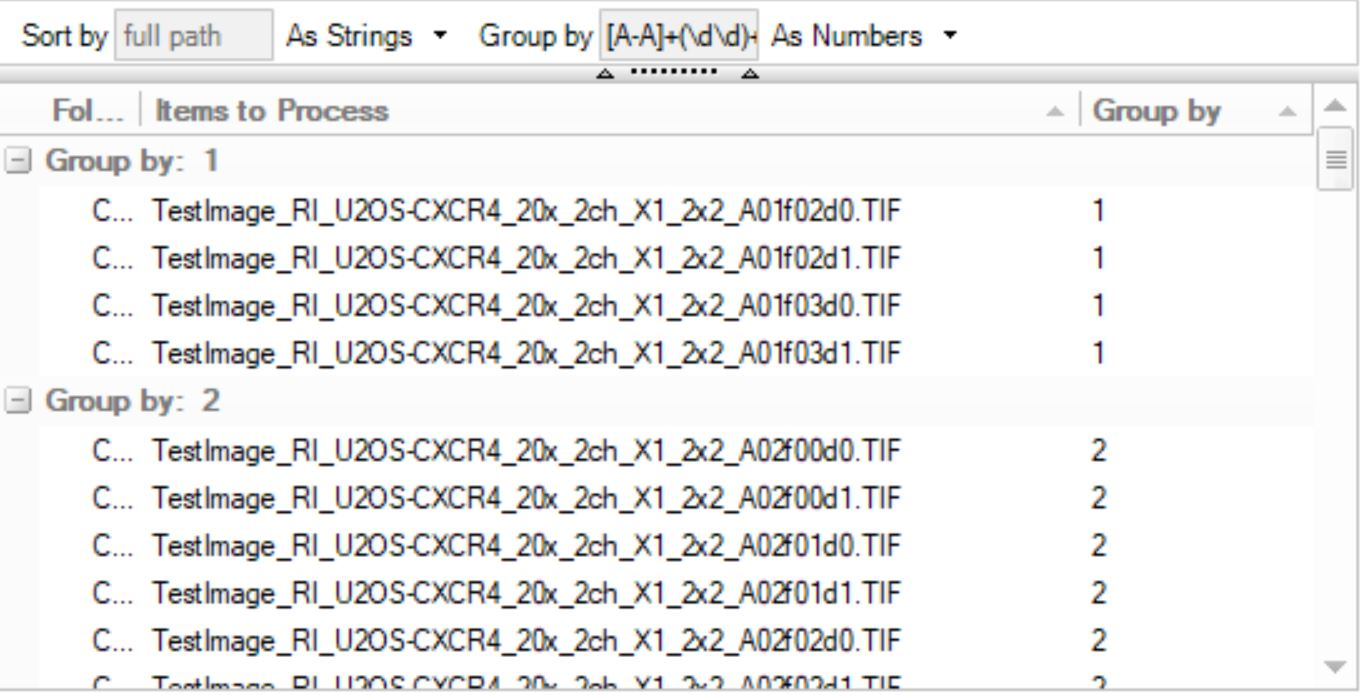
I'm trying to set up a Parent-Child batch analysis of a set of images. The images consist of multiple positions with every position having 2 channels (parent and child channel). Attached is the general naming of the files - d0 (parent) and d1 (child) are the two channels. A01 and f00 are identifiers of different positions.
I've tried grouping them accordingly using this syntax : [A-A]+(\d\d)+.+(\d\d). However it does not separate images with different numbers after 'f'. It should have 2 images per group.
Also, as I haven't been able to try this, would this even be the way to batch process images for the parent-child app?
Thank you,
Henri

0
Best Answer
-
BTW, the pattern that groups channel images of the same location would be:
Test(.*?)d
it would group by the part of file name that starts with "Test" (it could be any part of the image name) and ends with "d":
In my example below I used Plate(.*?)d
Yuri0
Answers
-
Hi Henri,
It can be tricky to group images in the folder the way you want. Do you have "scanptotocol" file? If yes, then you can open the whole folder as an image set (multi-well, multi XY, 2 channels) and process it using ImageSetAnalyzer app. Will it work for you?
Yuri
0 -
Thanks, Yuri.
Could you explain why it's difficult? It's recognising the 'A01', so it should be fine for 'f01'?!
I don't have a .scanprotocol file for this set of data, unfortunately. I tried copying the scanprotocol file from another set and change the name to match these images. However, images were displayed overexposed (which they are not) and counting options were greyed out. 0
0 -
Henri,
Regarding the "overexposed" images in the image set: opening images as "image set" (with scanprotocol) should not change intensity of the images. Adjusting the LUT or applying Best-Fit should make them look right. If it doesn't help, can you please send me 2 images (d0 and d1) and I will check them?
Also, the Count is disabled when Color Composite mode is active, you should switch it off.
Yuri0 -
Thanks Yuri!
Using Plate(.*?)d works perfectly! Although now I keep getting an error 'No Parent Objects'. It's worked once, but ever since I keep getting this error. Do you have a suggestion as to why? I know I have objects in the image, as I have set up my settings on it. It even showed me the number of objects during set up.
Also, what would happen if there ever was an image without parent objects? It seems to pause the whole batch process.
UPDATE: I realised you have to enable 'display documents'. It's working now.
Regarding you second post:
The scanprotocol file was from a different microscope, which had a lower bit-depth. I think this is causing the problem, although I changed the bit-depth value in the scanprotocol file. Your first post seems to work, but I'd be curious if the scanprotocol could be adjusted (please see attached).0
Categories
- All Categories
- 963 Image-Pro v9 and higher
- 9 Image-Pro FAQs
- 18 Image-Pro Download & Install
- 449 Image-Pro General Discussions
- 487 Image-Pro Automation (Macros, Apps, Reports)
- 20 AutoQuant Deconvolution
- 2 AutoQuant Download & Install
- 18 AutoQuant General Discussions
- 195 Image-Pro Plus v7 and lower
- 3 Image-Pro Plus Download & Install
- 106 Image-Pro Plus General Discussions
- 86 Image-Pro Plus Automation with Macros
- 19 Legacy Products
- 16 Image-Pro Premier 3D General Discussions
- 26 Image-Pro Insight General Discussions
